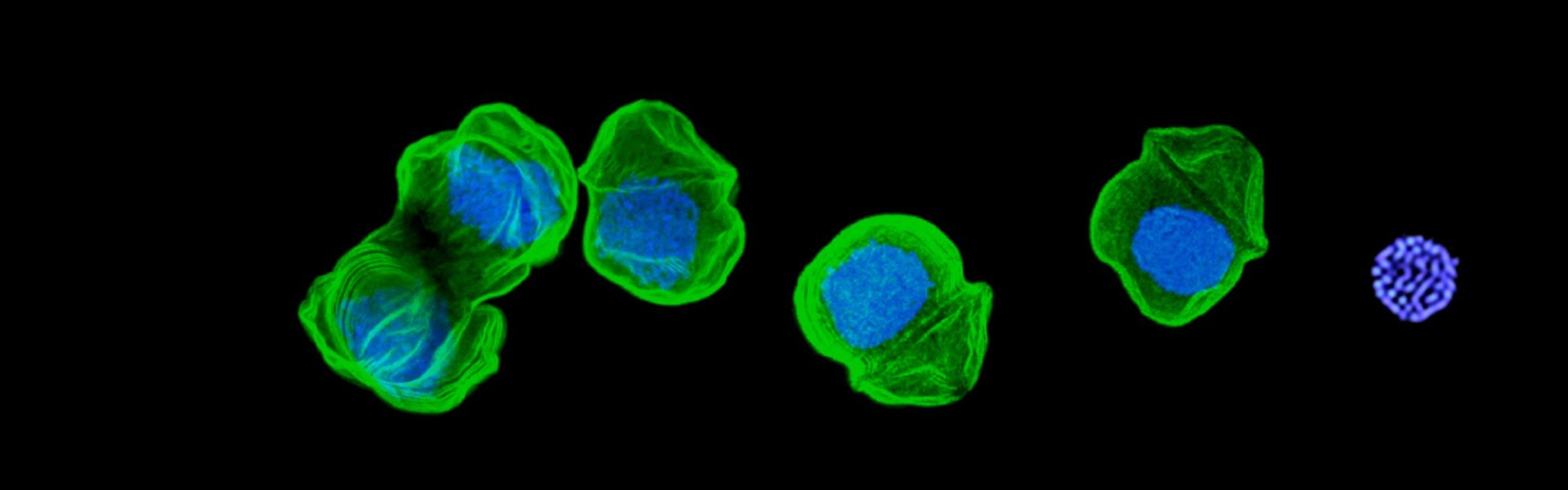
chromatin without histones and genes without stops
Eukaryotic nuclei define our difference from prokaryotes, and the use of histone proteins as DNA packing and genome regulation agents was a seminal invention. So profound was this innovation, that all eukaryotes continue to use it—all except dinoflagellates. We have shown that early in dinoflagellate evolution histones were largely abandoned and no longer play a role in bulk DNA packing. In its place, a new major nucleoprotein has been gained (DVNP), acquired from an algal virus. We are now studying genome architecture and management in this novel chomatin model, and the role and properties of DVNP. These studies are informing us of radical changes possible in nuclear organisation, and possible models for alternative chromatin systems that might be more broadly present in eukaryotes.
Eukaryotes also possess genetic material in their mitochondria and plastids, relicts of the endosymbionts’ own genomes. The Waller lab is investigating evolutionary processes that continue to shape these organelle genomes also. In dinoflagellate mitochondria, again remarkable change is seen. Since diverging from their sister taxa Apicomplexa, these genomes have lost conventional translation start and stop signals, acquired a novel mRNA trans-splicing process, and the genomes have been massively amplified and become highly recombinatorial, despite encoding only three protein genes. Moreover, extensive RNA editing has been acquired in these mitochondria, and this process is even imposed upon recently gained dinoflagellate endosymbionts.
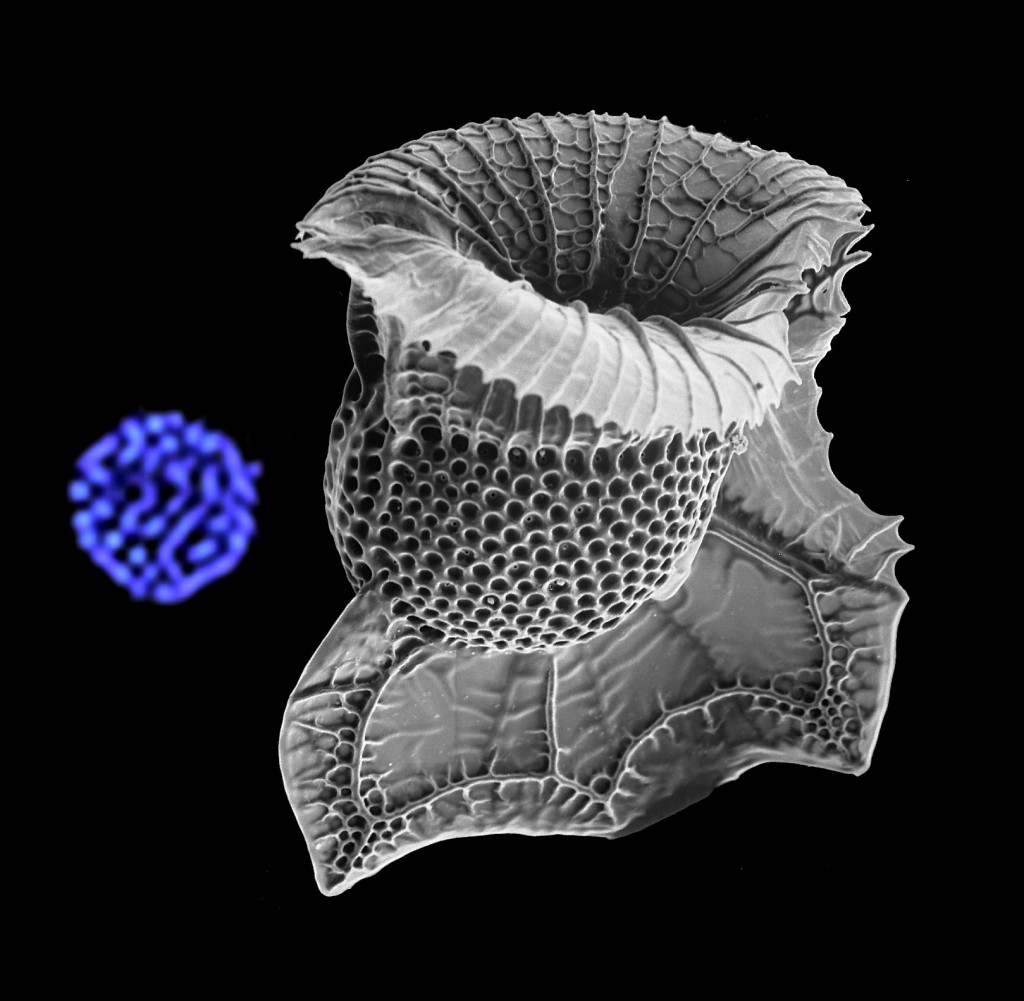
Dinoflagellate Ornithocercus and a dinokaryon confocal optical section showing condensed chromosomes in blue.
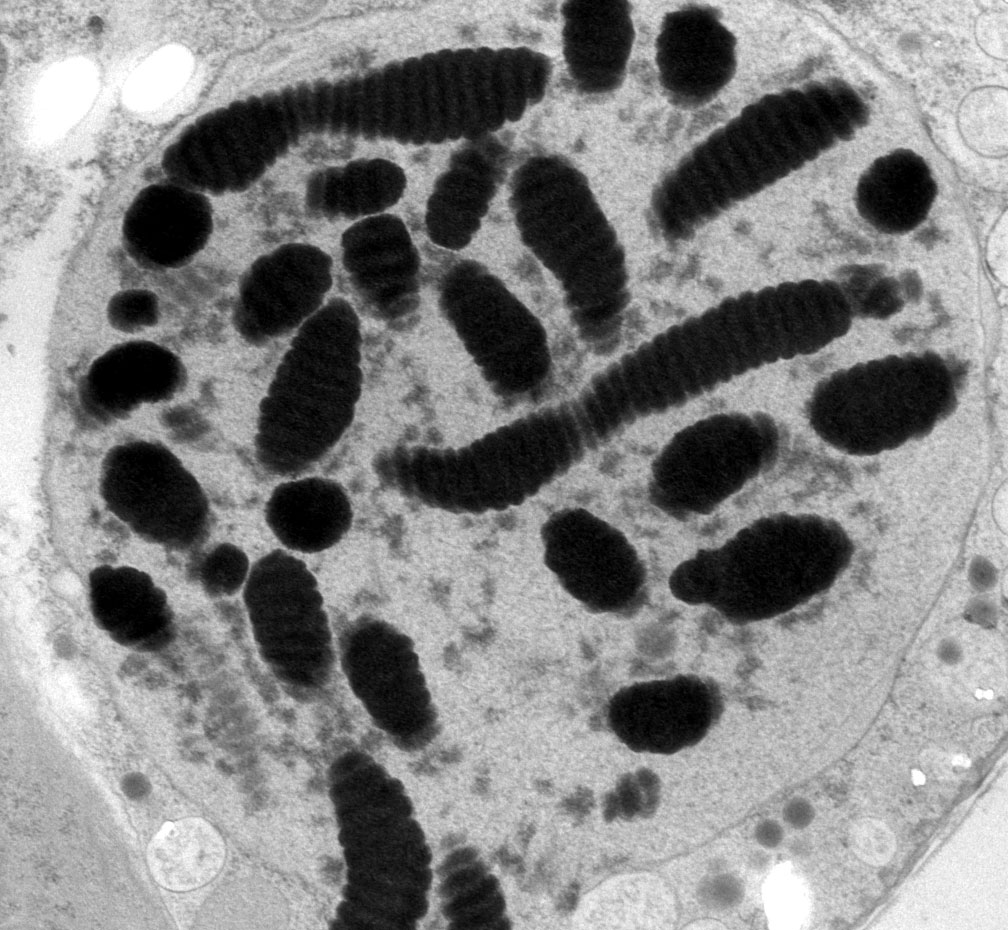
Thin section of the permanently condensed, highly structured, chromosomes of the "dinokaryon"
Selected publications:
- Jackson, C.J. et al (2013) A tertiary plastid gains RNA editing in its new host. Mol. Biol. Evol. 30(4):788-92
- Jackson, C.J. et al (2013) A widespread and unusual RNA trans-splicing type in dinoflagellate mitochondria. PLOS ONE 8(2): e56777
- Gornik, S.G. et al (2012) Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 22(24): 2303-12
- Jackson, C.J. et al (2012) The mitochondrial genome and transcriptome of the basal dinoflagellate Hematodinium sp.: character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biol. Evol. 4(1):59-72
- Burger, G. et al (2012) Unusual mitochondrial genomes and genes. In Organelle Genetics (ed Charles Bullerwell) Springer, New York, USA. pp 41-77
- Waller, R.F. et al (2009) Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. BioEssays 31(2):237-45
- Jackson, C.J. et al (2007) Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biology 5:4